Germline Genomics Analysis of Tissue-Cultured Plants
Phylogenies reflect natural history, and understanding phylogenetic relationships among species is fundamental to understanding evolution and biodiversity in biology. Lifeasible provides phylogenetic tree construction, tree examination, species divergence time estimation, and adaptive evolution analysis of tissue culture plants and other germline genome analysis services.
What is a Germline Genome?
Germline genome analysis is the use of computational biology methods to study phylogenetic relationships among species based on genome or transcriptome sequence data and to perform species and molecular evolutionary analyses based on phylogenetic trees. A phylogenetic tree is a hypothesis that describes the evolutionary relationships of a group of organisms. Phylogenetic trees can be constructed using morphological (body shape), biochemical, behavioral, or molecular traits. In constructing a phylogenetic tree, researchers group species according to the derived traits they share (traits that differ from their ancestors). Phylogenetic trees can also be constructed by comparing the sequences of genes or proteins. Closely related species usually have little or no sequence differences, while less related species tend to have large differences between them.

Accurate phylogenetic trees are the basis for researchers to understand important transitions in evolution and are key to inferring the origin of new genes, detecting molecular adaptations, understanding the evolution of morphological traits, and reconstructing changes in the number of recently diverged species. Despite the increasing abundance of data and powerful analytical methods, many challenges to reliable tree construction remain.
What We Offer
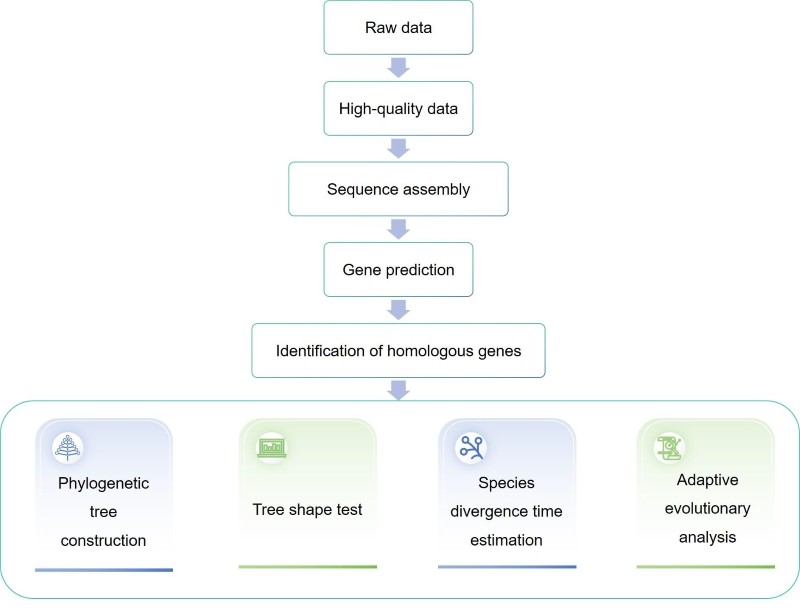
Transcriptome sequencing (RNA-seq) provides extensive transcriptional information from the genome. When genome-wide data are lacking, RNA-seq-based phylogenetic reconstruction is more efficient and cost-effective than traditional polymerase chain reaction-based or expressed sequence tag (EST) -based methods. We provide you with RNA-seq-based germline genome analysis, including identification of orthologous genes or proteins, multiple sequence alignment, selection of substitution models, and inference methods.
We provide you with phylogenetic processes for accurate tree construction, from careful data summarization, including immediate homologous gene identification and contamination avoidance through multiple sequence comparison, to a selection of tree reconstruction methods and replacement models to avoid systematic errors in phylogenetic reconstruction. For challenging phylogenies, especially deep phylogenies involving distant species, we choose likelihood-based methods and select appropriate models to accommodate heterogeneity during molecular evolution across loci, taxa, and time.
Technology route
Advantages
- Personalized research protocols and specialized bioinformatics analysis based on research objectives.
- Professional experimental team to provide highly accurate sequencing data.
- Experienced project team with in-depth analysis of difficult problems.
Sample Requirements
| Sample type |
Sample concentration |
Total sample volume |
Second-generation sequencing depth |
| DNA |
≥10 ng/μL |
≥1 μg |
≥30 × |
| Plant tissue |
- |
≥2 g |
Lifeasible has deeper insights into screening for immediate homologous genes, multiple sequence pairs, assessment of compositional heterogeneity and missing data, selection of evolutionary models, construction of phylogenetic trees, analysis of anomalous species, analysis of differences between species and gene trees, and estimation of the time of species divergence. Please feel free to contact us for more information about the germline genomics analysis of tissue-cultured plants.
You want to sign a confidentiality agreement.
You have a specific plant species for your experimental needs.
You have a reliable and relevant cooperation project to discuss.
You are very interested in our project or have any questions.
You need an updated and detailed quotation.
For research or industrial use.