Development of SSR Molecular Markers for Tissue Culture Plants
Simple sequence repeats (SSR) markers are widely used in genetic mapping, population genetics, fingerprinting, phylogeny, and other research fields due to their rich polymorphism, reproducibility, and co-occurrence. Lifeasible provides customers with PCR product scanning and data analysis and designs SSR molecular marker development experiments for customers' specific tissue culture plant samples. Including the selection of fluorescent markers, primer synthesis, fluorescent molecular weight marker selection, etc., and completing the SSR & STR typing service for specific samples.
What are Molecular Markers?
Conventional plant genetic breeding has played a great role in promoting the development of agriculture but suffers from the defects of long breeding cycles and low efficiency of genetic improvement practice. Molecular design breeding based on molecular biology, genomics, bioinformatics, and other technologies has the incomparable advantages of conventional breeding. Functional genomics research results can be quickly converted into field crop varieties, thus creating huge economic benefits and ensuring sustainable development of agriculture. Molecular marker is a very important concept in breeding, which is a direct reflection of genetic polymorphism at the DNA level, and has been widely used in genetic breeding, gene localization, species kinship identification, and other aspects.
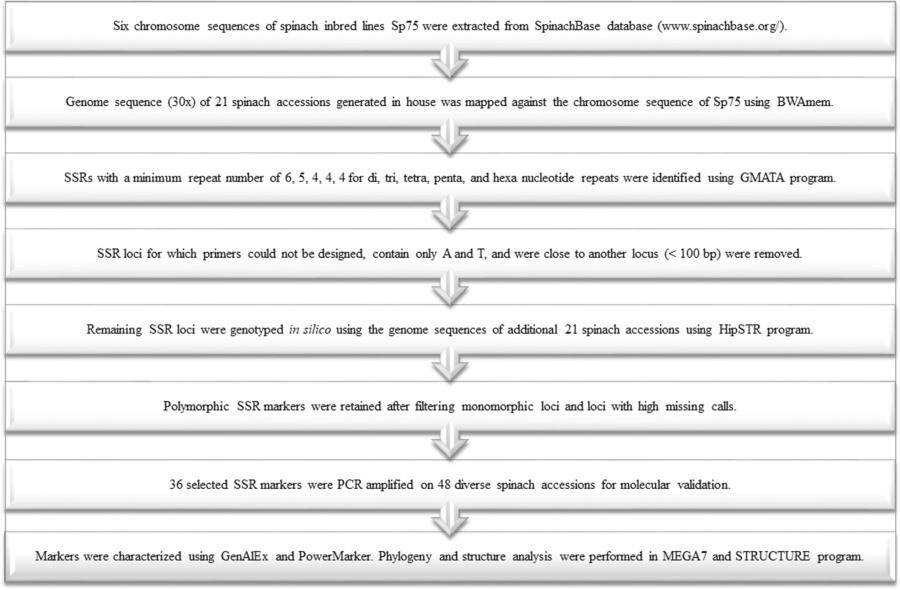 Figure 1. Outline of the approach used to discover SSR loci from the genome sequence, the stepwise procedure employed to identify a large set of polymorphic loci, and genetic characterization and diversity analysis. (Bhattarai, G., et al, 2021)
Figure 1. Outline of the approach used to discover SSR loci from the genome sequence, the stepwise procedure employed to identify a large set of polymorphic loci, and genetic characterization and diversity analysis. (Bhattarai, G., et al, 2021)
SSR refers to tandem repeat sequences consisting of 1-6 nucleotides in the genome, which are widely and relatively uniformly distributed in various eukaryotic genomes. Polymorphisms in the length of SSR gene fragments can be used as molecular markers, and through the analysis of DNA polymorphisms such as SSR markers in the genome of a sample, the regulation and differences of the sample DNA sequences in genetic traits can be obtained. Because SSR markers are rich in polymorphisms, stable, usually dominant consistent with Mendelian inheritance laws, and informative, they are widely used in target gene calibration, fingerprinting, and other research. With the emergence of next-generation sequencing (NGS), species genome information and SSR sequence information can be obtained in large quantities in a short time.
What We Offer
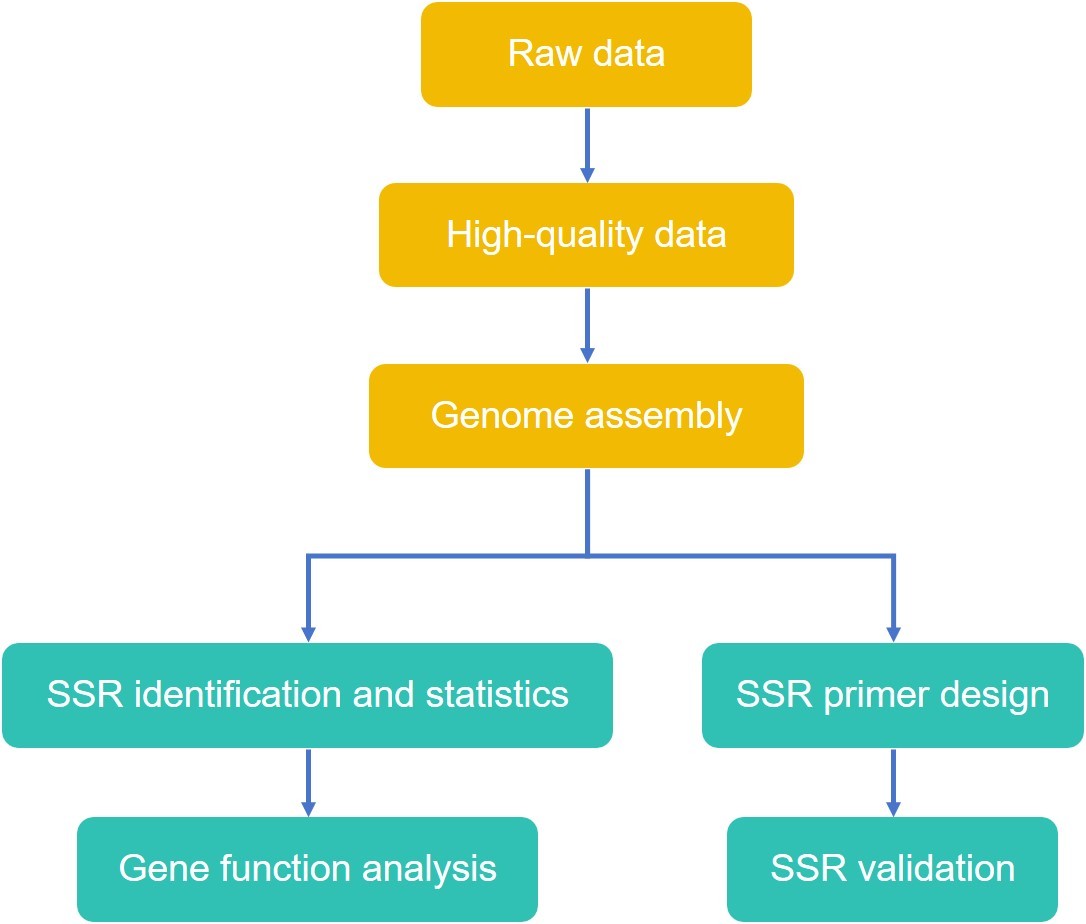
While applying traditional SSR marker development methods, we have been exploring technical services that can simplify the technical process, improve efficiency, and reduce development costs. We offer a variety of SSR marker development methods with enrichment steps.
- Randomly amplified polymorphic DNA (RAPD) based development strategy. Repeat sequences can be effectively enriched by attaching RAPD random primers to SSRs as PCR amplification primers or by Southern hybridizing RAPD amplification product strips with SSR probes.
- SSR isolation strategy based on ISSR-PCR. The separation method based on ISSR-PCR avoids the randomness of SSR enrichment and has a clearer purpose and stronger enrichment ability. It avoids the construction and screening of libraries, thus greatly shortening the time and reducing the research cost.
- Separation strategy based on primer extension. The initial single-stranded genomic library was transferred into a dut-ung- E. coli strain for proliferation, and then SSRs were used as primers to synthesize double-stranded vectors and insertion fragment DNAs, and further transfected with ung+ E. coli. Single-stranded DNA without SSRs was difficult to transfer and enrich, while double-stranded DNA containing SSR motifs was enriched.
- SSR isolation was performed using selective hybridization.
- Sequence tag libraries were created to obtain SSR primer sequences.
General technical routes
Advantages
- According to the research purpose, we develop a personalized research plan and specialized bioinformatics analysis.
- Professional laboratory team to provide highly accurate sequencing data.
- Experienced project team with in-depth analysis of difficult problems.
Sample Requirements
| Sample type |
Sample concentration |
Total sample volume |
Second-generation sequencing depth |
| DNA |
≥10 ng/μL |
≥1 μg |
≥100 × |
| Plant tissue |
- |
≥5 g |
Applications
- Genetic mapping
- Germplasm identification
- Genetic diversity analysis
- Gene localization
- Quantitative trait loci (QTL) analysis
Please feel free to contact us for more information about the development of SSR molecular markers for tissue culture plants.
Reference
-
Bhattarai, G., et al. (2021). “Genome-wide simple sequence repeats (SSR) markers discovered from whole-genome sequence comparisons of multiple spinach accessions. " Scientific Reports 11, 9999.
You want to sign a confidentiality agreement.
You have a specific plant species for your experimental needs.
You have a reliable and relevant cooperation project to discuss.
You are very interested in our project or have any questions.
You need an updated and detailed quotation.
For research or industrial use.

 Figure 1. Outline of the approach used to discover SSR loci from the genome sequence, the stepwise procedure employed to identify a large set of polymorphic loci, and genetic characterization and diversity analysis. (Bhattarai, G., et al, 2021)
Figure 1. Outline of the approach used to discover SSR loci from the genome sequence, the stepwise procedure employed to identify a large set of polymorphic loci, and genetic characterization and diversity analysis. (Bhattarai, G., et al, 2021)