Genome De Novo Sequencing of Tissue Culture Plants
Without relying on reference genomes, Lifeasible can obtain the entire genome sequence information in the nucleus of tissue culture plant cells through next-generation sequencing technology. Based on this information, we can help you study the evolutionary relationships, genome evolution, and molecular evolution of the species. At the same time, we build a comprehensive research platform for the species to ensure the subsequent functional gene mining, analysis, and experimental validation needs.
What is Genome De Novo Sequencing?
Genome de novo sequencing, or genome de novo sequencing, refers to sequencing the whole genome sequence of a species without relying on known genome sequence information. The sequenced sequence is then spliced and assembled using bioinformatics to obtain a whole-genome sequence map of the species. The construction of the whole genome sequence atlas will comprehensively deepen the understanding of the evolutionary process of the origin of the species as well as its adaptation to a specific environment. It will play a great role in the future discovery of new genes in this species and the improvement of the species, and build an efficient platform for genomics research, providing DNA sequence information for subsequent gene mining and functional verification.
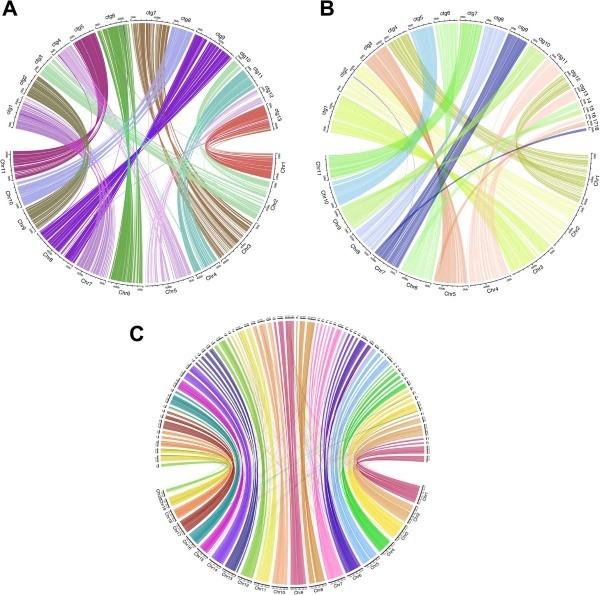 Figure 1. Chord diagrams of E. camaldulensis, E. rudis, and A. hypogaea de novo assemblies mapped against a reference genome. (Driguez, P., et al, 2021)
Figure 1. Chord diagrams of E. camaldulensis, E. rudis, and A. hypogaea de novo assemblies mapped against a reference genome. (Driguez, P., et al, 2021)
What We Offer
- Genome survey: evaluate genome GC content, duplication, heterozygosity, and estimate genome size.
- Genome splicing statistics: splicing statistics, including raw data statistics, sequencing depth, Contig N50, Scaffold N50, and genomic GC content.
- Genome annotation and gene function classification: including gene prediction, function annotation, ncRNA annotation, repetitive sequence analysis, as well as GO classification and KEGG pathway analysis.
- Comparative genomic and evolutionary analyses: nucleotide level covariance analysis, amino acid level covariance analysis, gene cluster analysis, and evolutionary relationships.
Technology route
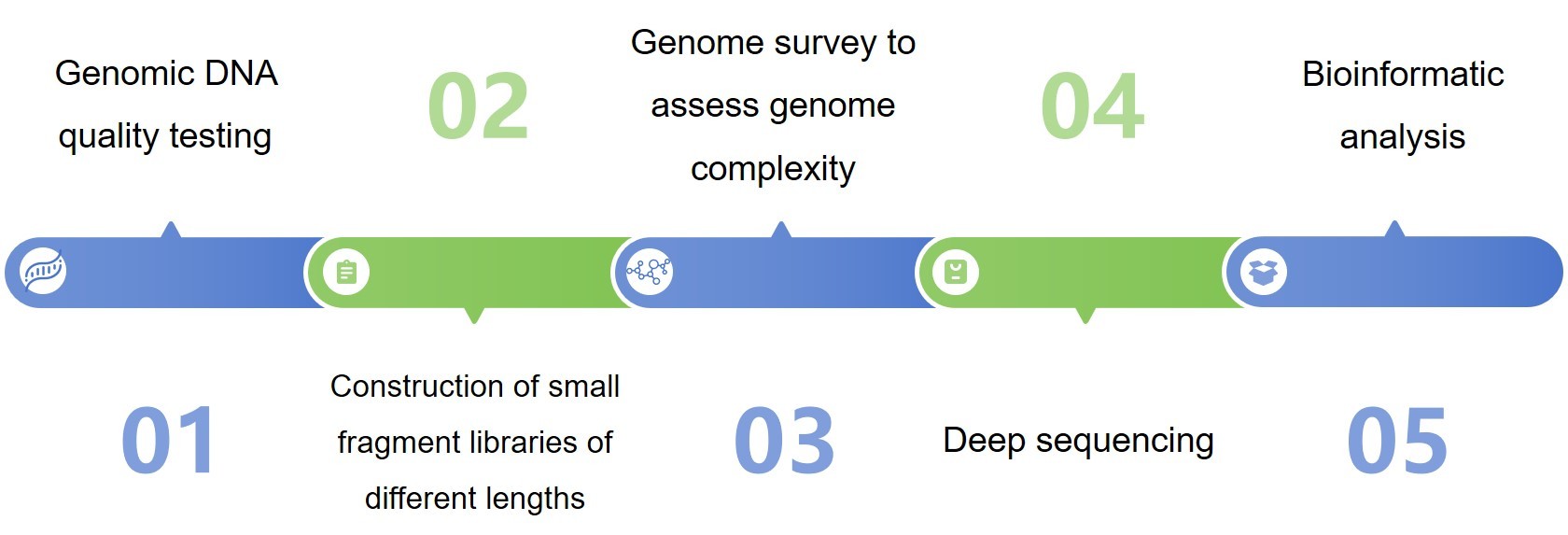
Content of analysis
|
Standard information analysis |
Advanced information analysis |
- Data processing: removal of splice sequences, contaminated sequences, etc.
- Genome assembly (third generation data): raw data statistics, sequencing depth analysis, coverage statistics, GC content analysis, etc.
- Genome correction (second generation data): genome comparison and correction.
- Sequence integrity assessment: CEGMA assessment, BUSCO assessment, EST/transcript sequence assessment.
- Genome annotation: gene prediction, gene function annotation, repetitive sequence analysis, non-coding RNA annotation.
- Comparative genomics and evolutionary analysis: species phylogenetic tree construction, gene family identification, gene covariance analysis, etc.
|
- K-mer analysis and genome size estimation.
- Heterozygosity estimation.
- Preliminary assembly.
- GC-Depth distribution analysis.
|
Sample Requirements
- Total amount of DNA sample: Small fragment library sample preparation requires a total sample greater than 1 μg. Mate-pair library (2 kb, 5 kb, 10 kb) sample preparation requires 20-50 μg.
- Sample concentration and purity: >200 ng/μl, OD260/280 between 1.8 and 2.0, no contamination by protein, RNA, or visible impurities.
- Sample storage: Please store the sample in dry powder, alcohol, TE buffer, or ultrapure water, and indicate this in the sample information sheet.
- Sample quality: genome integrity, no degradation, clear electrophoretic main bands, no dispersion.
Please feel free to contact us for more information about the genome de novo sequencing of tissue culture plants.
Reference
- Driguez, P., et al. (2021). “LeafGo: Leaf to Genome, a quick workflow to produce high-quality de novo plant genomes using long-read sequencing technology. " Genome Biology 22, 256.
You want to sign a confidentiality agreement.
You have a specific plant species for your experimental needs.
You have a reliable and relevant cooperation project to discuss.
You are very interested in our project or have any questions.
You need an updated and detailed quotation.
For research or industrial use.

 Figure 1. Chord diagrams of E. camaldulensis, E. rudis, and A. hypogaea de novo assemblies mapped against a reference genome. (Driguez, P., et al, 2021)
Figure 1. Chord diagrams of E. camaldulensis, E. rudis, and A. hypogaea de novo assemblies mapped against a reference genome. (Driguez, P., et al, 2021)